Knocking-out all 28 genes responsible for plant-specific protein glycosylation for LenioBio
In 2019, LenioBio approached us with a challenge for our CRISPR gene editing workflow. LenioBio provides a cell-free protein production platform based on a lysate from the tobacco BY-2 cell line: ALiCE®.
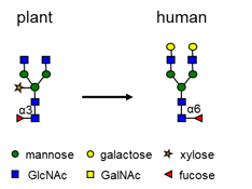
For LenioBio’s cell line to produce proteins that are better suitable for pharmaceutical applications, plant-specific sugars should be removed to eliminate the risk of a human immune response to such plant-like proteins.
Their request to us was to knock-out in a single cell 28 genes responsible for plant-specific protein glycosylation with fucose and xylose: fucosyltransferases (FucTs) and xylosyltransferases (XylTs).
What we did
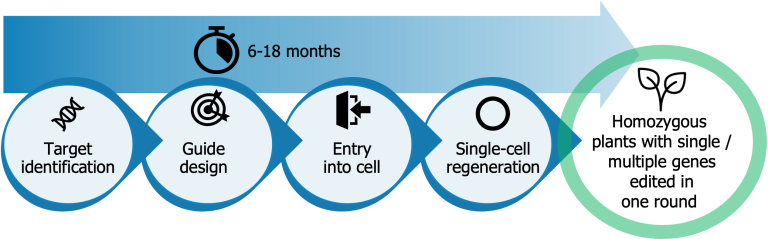
The tobacco BY-2 cell line is a tetraploid and has been cultured over the last 50 years. In this project, we made use of our proprietary TiGER (Target identification, Guide design, Entry into the cell, Regeneration) workflow, which overcomes CRISPR bottlenecks for a wide range of species.
Target identification
Guide design
Entry into the cell
Regeneration
During the first phase, Target identification, we identified 7 relevant targets responsible for plant-specific protein glycosylation (in total 28 alleles). Next, in the Guide design phase, we grouped gene targets based on homology and designed, tested and selected the best guides per gene group. In the next phase (Entry into the cell), we used PEG-based transfection of protoplasts and the MAD7 CRISPR endonuclease, which in HRB’s hands can achieve high editing efficiencies of 40-60% in protoplasts (reducing costs and effort for edit screening). In the final phase, Regeneration, we applied our proprietary single-cell regeneration protocol and performed a PCR and antibody-based screening to identify and confirm the mutants we were looking for.
What we achieved
In a period of 18 months, we developed the required protocols and delivered Tobacco BY-2 cell lines lacking plant-specific glycosylation for LenioBio’s ALiCE®. We achieved this by using our proprietary, transgene free CRISPR editing workflow. After further testing, LenioBio confirmed the multiple knock-out, which performs very well in culture and results in production of proteins lacking plant-specific Xyl or Fuc moieties for ALiCE®.
What they said


How does our TiGER workflow address common CRISPR bottlenecks?
For a wide range of species, our TiGER workflow overcomes CRISPR bottlenecks to rapidly deliver top-quality products that can be directly introduced into the market development pipeline. In this way, we tackle the most common challenges associated with CRISPR gene editing.
Our proprietary TiGER workflow yields a genetically uniform edited organism in 6-18 months, offering benefits compared to traditional approaches:
- Transgene-free delivery protocols without the use of foreign DNA, which limits regulatory issues;
- MAD7 nuclease, a commercially more attractive option than Cas9, with high editing efficiencies to minimize costs for edit screening;
- Proprietary guide design software to target single or multiple genes in one editing round while minimizing chances of off-target edits;
- Single-cell protoplast regeneration to avoid edited chimeras. Even for recalcitrant species difficult to handle in vitro, we have achieved CRISPR active delivery in intact cells using nanoparticle-based transfection methods.
By applying our workflow, you can speed up the development of plant breeding, even in comparison to other CRISPR approaches.
Interested in learning more or have a general inquiry?
Read about our proprietary CRISPR workflow in the whitepaper
Our TiGER workflow enables molecular breeding in an end-to-end fashion, providing innovative solutions to the typical barriers encountered in CRISPR breeding projects. Read about our step by step approach by downloading our whitepaper.